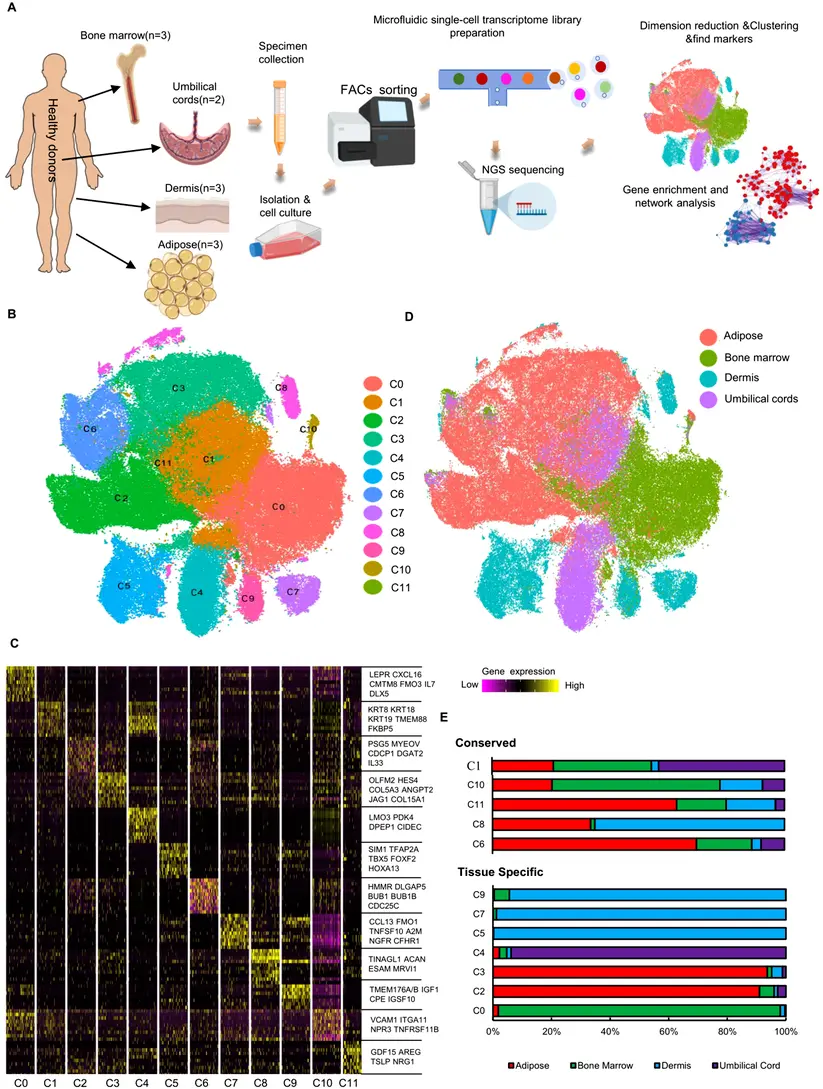
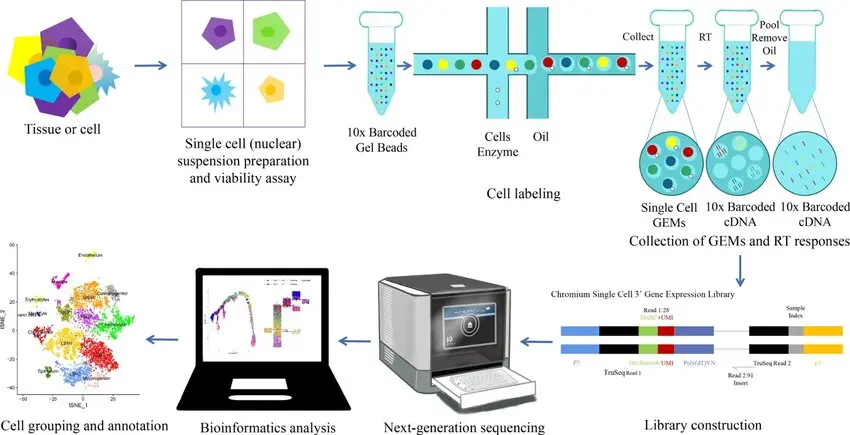
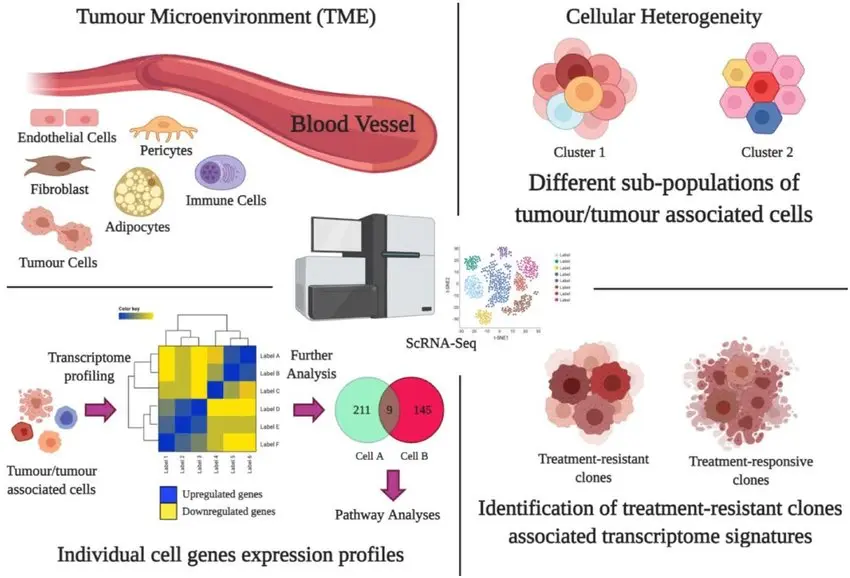
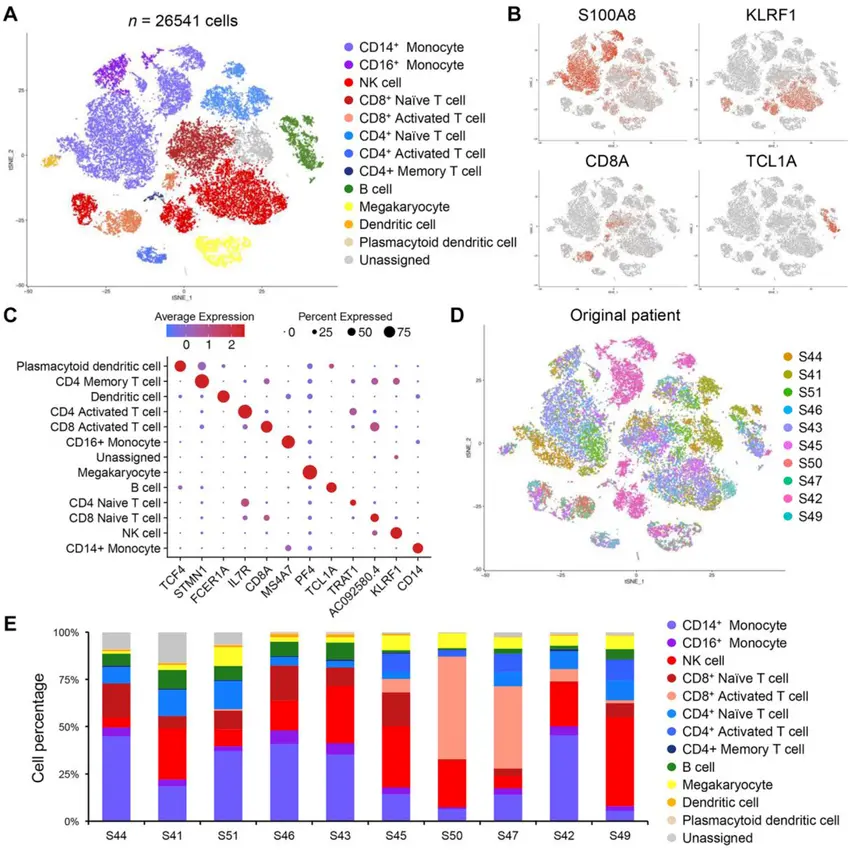
One technological innovation that has revolutionized contemporary biology and biomedical research is single-cell RNA sequencing, or scRNA-seq. scRNA-seq allows for the study of individual cells by capturing their distinct molecular profiles, variations, and behaviors, in contrast to traditional RNA sequencing (RNA-seq), which measures gene expression across bulk populations of thousands or millions of cells. This method is revolutionizing our understanding of cellular complexity in health, disease, development, and evolution.
Why Single-Cell RNA Sequencing?
Biological tissues are not uniform; they are made up of numerous cell types, each of which has a specific function. Depending on environmental cues or disease conditions, cells can exist in different functional states, including active, resting, dividing, or dying, even within the same tissue. Traditional bulk RNA sequencing masks these differences, providing an average gene expression profile that overlooks critical heterogeneity.
This restriction is removed by scRNA-seq, which analyzes gene expression per cell. Discovering hidden cell types, monitoring cellular development, and comprehending dynamic biological processes like immune responses or tumor progression all depend on the capacity to listen to the molecular language of thousands to millions of individual cells operating in parallel.
How Does scRNA-seq Work?
The process of single-cell RNA sequencing includes several precise and innovative steps:
Cell Isolation
Individual cells are separated from a tissue or fluid sample using microfluidics, droplet-based systems, or manual techniques like fluorescence-activated cell sorting (FACS). This step is critical to ensure that each RNA transcript detected comes from only one cell.
RNA Capture and Conversion
The RNA molecules (messengers from DNA) inside each isolated cell are captured and reverse-transcribed into complementary DNA (cDNA). Unique molecular barcodes are added to tag the cDNA from each cell, allowing the data to trace every sequence back to its original cell.
Library Preparation and Sequencing
The barcoded cDNA libraries are amplified and prepared for sequencing, usually performed on platforms like Illumina or Oxford Nanopore. Massive parallel sequencing generates millions or billions of short reads that represent the transcriptome of each individual cell.
Data Analysis and Interpretation
Specialized bioinformatics pipelines cluster cells based on gene expression similarities, detect rare cell types, identify new cell states, and reconstruct developmental pathways. Data visualization tools such as t-SNE or UMAP help researchers see how different cells relate to one another in expression space.
Applications of scRNA-seq in Modern Science
1. Understanding Developmental Biology
scRNA-seq has provided revolutionary insights into embryonic development by mapping the progression of stem cells into specialized tissues. Researchers can now track how a single fertilized egg differentiates into neurons, muscle cells, or blood cells revealing the genetic programs guiding these processes.
2. Cancer Research and Tumor Heterogeneity
Cancer is not a single disease but a complex mixture of cell populations with varying genetic mutations. scRNA-seq allows the detection of resistant clones, metastatic cells, and immune infiltrates within tumors, offering clues for better-targeted therapies and personalized medicine.
3. Immunology and Infectious Disease
By profiling individual immune cells (such as T cells or macrophages), scRNA-seq reveals how the immune system responds to infections, vaccines, or autoimmune triggers.
4. Neuroscience
The brain contains an astonishing diversity of cell types. scRNA-seq is helping neuroscientists build complete cell atlases of the brain, discovering new neuronal subtypes, and understanding diseases like Alzheimer’s or Parkinson’s at the cellular level.
Advantages of scRNA-seq Technology
✔ Uncovers rare or hidden cell types that cannot be detected by bulk sequencing.
✔ Provides high-resolution views of cellular diversity and functional states.
✔ Tracks dynamic changes during development, disease progression, or response to therapy.
✔ Enables construction of cellular atlases, offering a reference for future research and diagnostics.
The Future of scRNA-seq
The future of single-cell RNA sequencing is exciting. Emerging trends include:
- Multi-omics integration, combining RNA, DNA, protein, and epigenetic data from the same cell.
- Spatial transcriptomics, preserving the location of each cell within tissues while sequencing their RNA.
- Real-time single-cell sequencing, for fast, in-situ analysis even in clinical or field settings.
These developments will push the boundaries of personalized medicine, regenerative therapies, and our fundamental understanding of life.
Single-Cell RNA Sequencing has become an indispensable tool in biological research. By revealing the molecular conversations happening inside individual cells, this technology is transforming how we understand health, disease, and the incredible complexity of living organisms. As scRNA-seq continues to evolve, it holds the promise of reshaping medicine, and biotechnology in the coming decades.
For further information
Here you can find a current, comprehensive article on this topic:
Single-cell RNA sequencing technologies and applications: A brief overview