GCAT-SEEK: Empowering the Next Generation Through NGS and Biotechnology Innovation
Next-Generation Sequencing (NGS) at the Core of Modern Biotechnology Education

About Us
GCAT-SEEK continues the tradition of making genomics part of education by focusing on the exciting world of Next-Generation Sequencing (NGS) and biotechnology. The goal is to help more people use these advanced tools to study DNA, RNA, and biological systems in research and learning.
What is Next-Generation Sequencing (NGS)?
Next-Generation Sequencing (NGS) represents a suite of high-throughput technologies that rapidly and simultaneously sequence millions to billions of DNA or RNA fragments. This advancement has revolutionized life sciences by dramatically increasing the speed, scale, and affordability of sequencing.

Why NGS Matters:
- Speed: Entire genomes can be sequenced in a matter of days or even hours.
- Scale: Massive amounts of data reveal subtle genetic differences, gene expression patterns, and epigenetic marks.
- Applications: From personalized medicine and disease diagnosis to evolutionary biology.
The NGS Workflow:
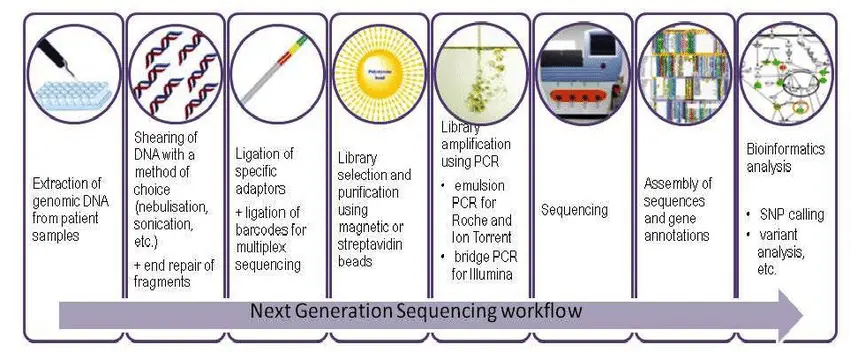
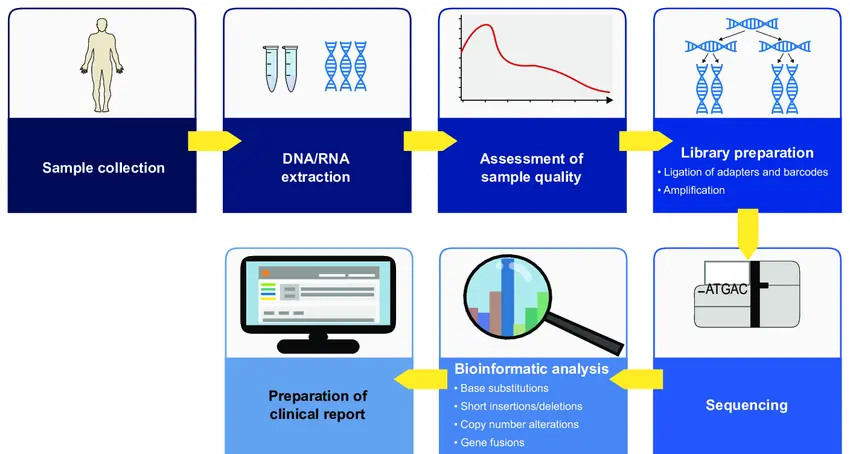
1. Sample Preparation
The process starts by carefully collecting biological material from humans or animals:
-
Sources of DNA or RNA:
- Blood samples
- Tissues (like skin, muscle, liver)
- Saliva or body fluids (for non-invasive sampling)
- Cells from biopsies or cultured cells
Extraction and Purification:
- DNA or RNA is extracted from these materials using laboratory kits or protocols.
- It is essential to get high-quality, pure nucleic acids—free of proteins, fats, or contaminants—since poor-quality material can ruin the sequencing process.
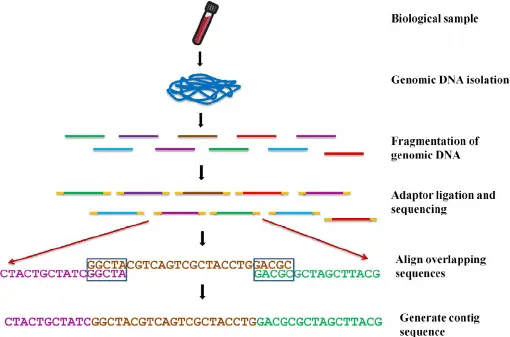
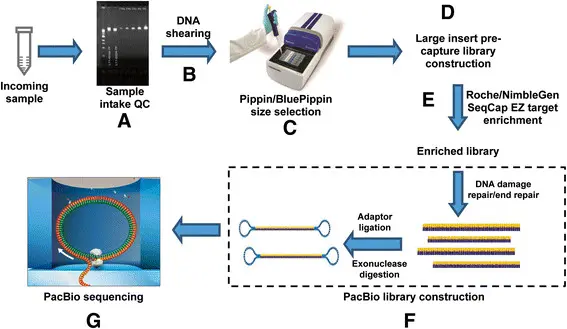
2. Library Construction
Before sequencing can happen, the DNA or RNA must be prepared:
-
Fragmentation:
The extracted DNA/RNA is broken into smaller pieces. This makes it easier for the sequencer to read them properly. -
Adapter Ligation:
Special short sequences of DNA (called "adapters") are attached to both ends of each fragment. -
Amplification:
The library is copied (via PCR) to make many copies, ensuring enough material is ready for sequencing.
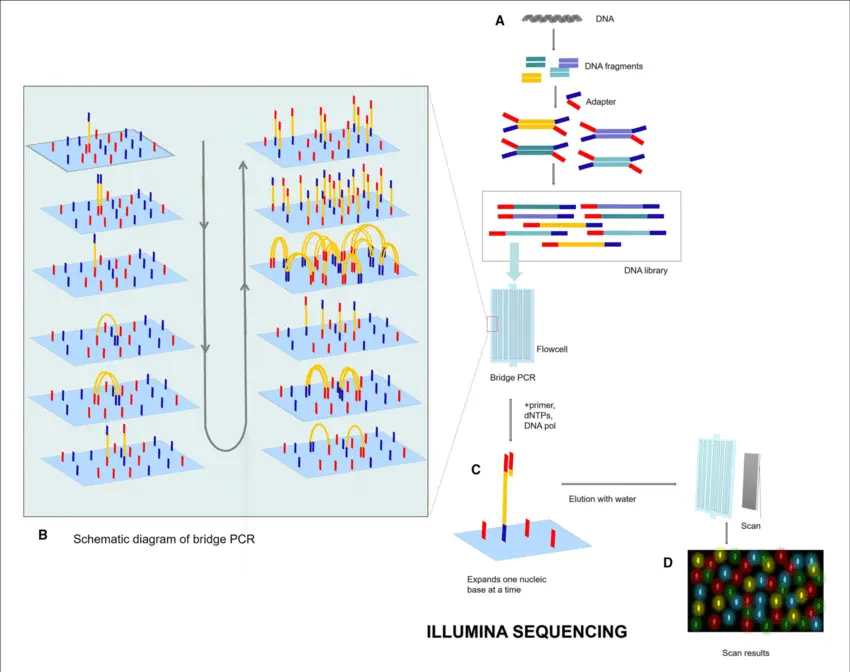
3. Sequencing Run
Now the prepared DNA/RNA library goes into a Next-Generation Sequencer (e.g., Illumina, Oxford Nanopore, PacBio):
-
Loading:
The library is placed onto a flow cell or chip in the sequencer. -
Sequencing:
The machine reads each fragment, one base at a time (A, T, C, G), generating millions or billions of short sequences (reads) simultaneously.
This process depends on the technology:- Sequencing by Synthesis (Illumina)
- Real-time long-read sequencing (Nanopore, PacBio)
4. Data Analysis
After sequencing, the real work begins on computers:
-
Quality Control (QC):
Remove poor-quality or contaminated reads to ensure reliable results. -
Alignment to Reference Genome:
Each read is compared to a known genome (e.g., human, mouse, or cow) to see where it fits. -
Variant Calling:
Detect differences like SNPs (single nucleotide polymorphisms), insertions, deletions, or larger structural changes. -
Gene Expression Analysis (for RNA-Seq):
Determine which genes are active and how strongly they are expressed under certain conditions (such as disease, stress, or treatment). -
Functional & Pathway Analysis:
Identify what biological functions or pathways are affected, and understand their role in health, development, or disease.
Types of Next-Generation Sequencing (NGS)
Next-Generation Sequencing (NGS) encompasses a variety of powerful technologies designed to address different biological questions. Each type of NGS serves a specific purpose, depending on the research goals and the biological material involved. Understanding these different methods is essential for students who wish to explore the modern world of genomics, biotechnology, and biomedical research.

Whole Genome Sequencing (WGS)
Whole Genome Sequencing (WGS) reads an organism’s complete DNA sequence from end to end. This method provides the most comprehensive genetic overview, capturing both coding and non-coding regions of the genome. WGS enables the discovery of new genes, the identification of mutations, the study of genome structure variations, and insights into evolutionary changes. It is commonly used in projects that require a full picture of an organism's genetic blueprint, whether in humans, animals, or microbes.
Whole Exome Sequencing (WES)
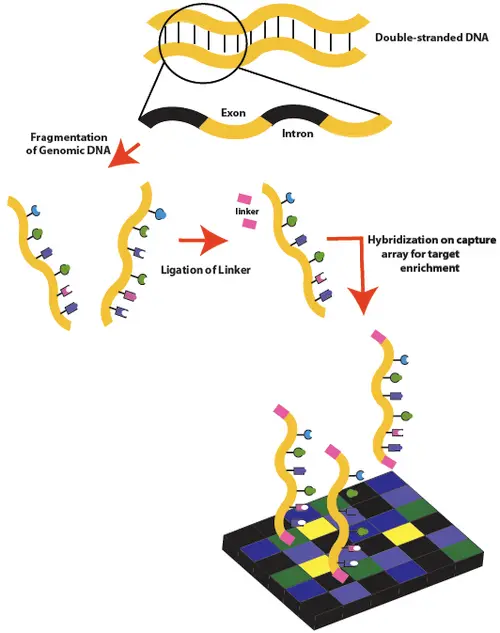
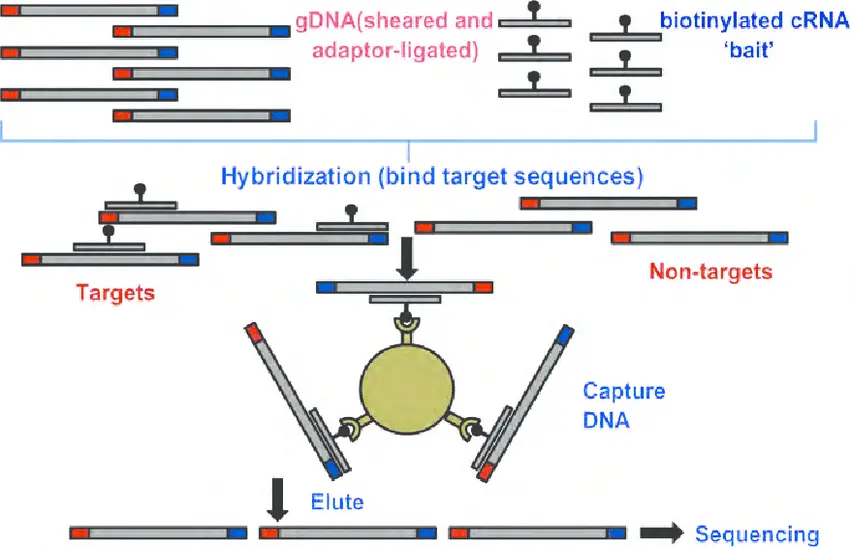
Whole Exome Sequencing (WES) focuses only on the exons — the protein-coding regions of the genome, which represent just 1–2% of total DNA but contain most mutations associated with traits and diseases. This approach is faster and more cost-effective than WGS while still delivering valuable information about gene function and variation. WES is widely applied in human and animal disease studies, such as identifying mutations responsible for inherited conditions or researching traits important for livestock improvement.
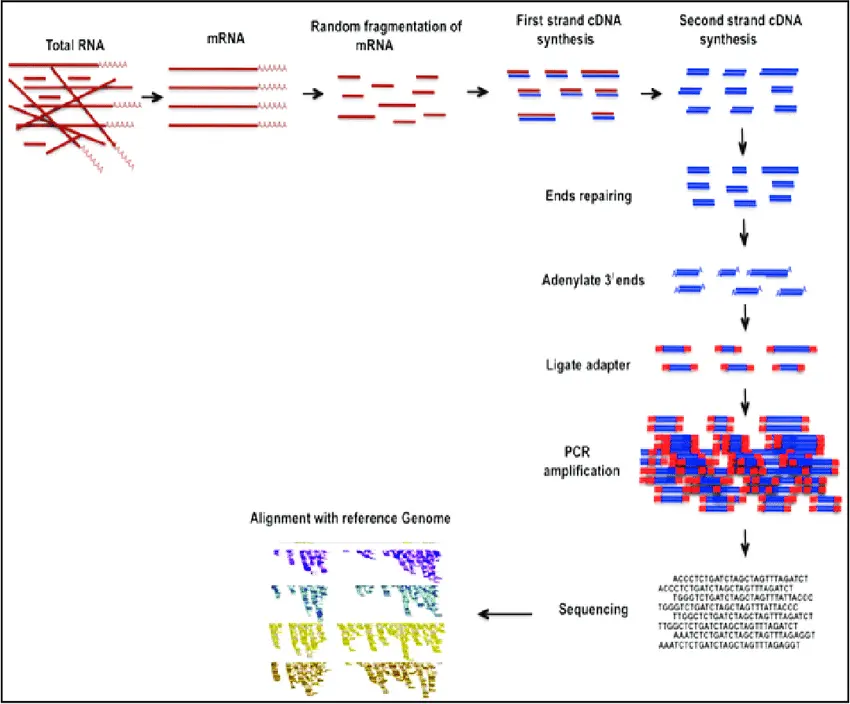
RNA Sequencing (RNA-Seq)
RNA Sequencing (RNA-Seq) is used to measure gene expression by sequencing RNA transcripts instead of DNA. This method reveals which genes are turned on or off in a cell, tissue, or organism under different conditions such as development stages, disease states, or environmental stress. RNA-Seq helps scientists and students understand how cells respond to treatments, how diseases alter gene activity, or how different tissues express unique sets of genes.
Targeted Sequencing (Amplicon Sequencing)
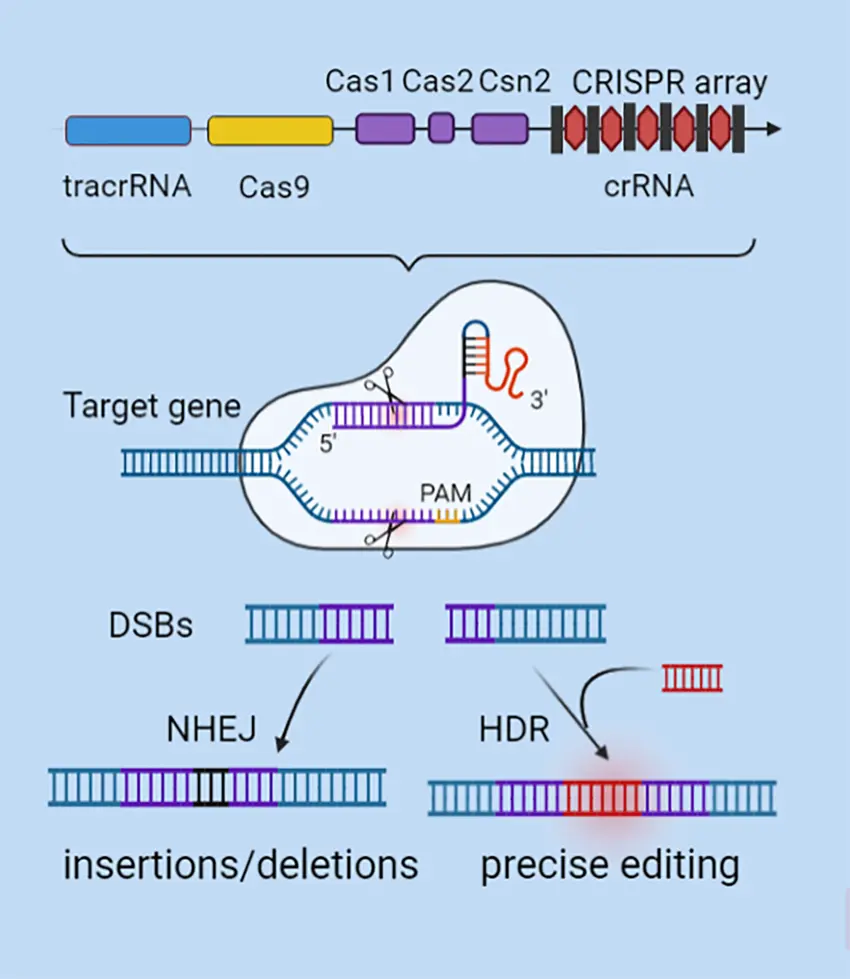
Targeted Sequencing focuses on specific regions of interest within the genome, such as a single gene or a panel of genes known to be involved in particular traits or diseases. This technique allows researchers to gather deep, highly accurate data from selected locations rather than sequencing the whole genome. Targeted sequencing is ideal for mutation screening, studying microbial diversity in samples, or validating gene editing (such as CRISPR) outcomes.
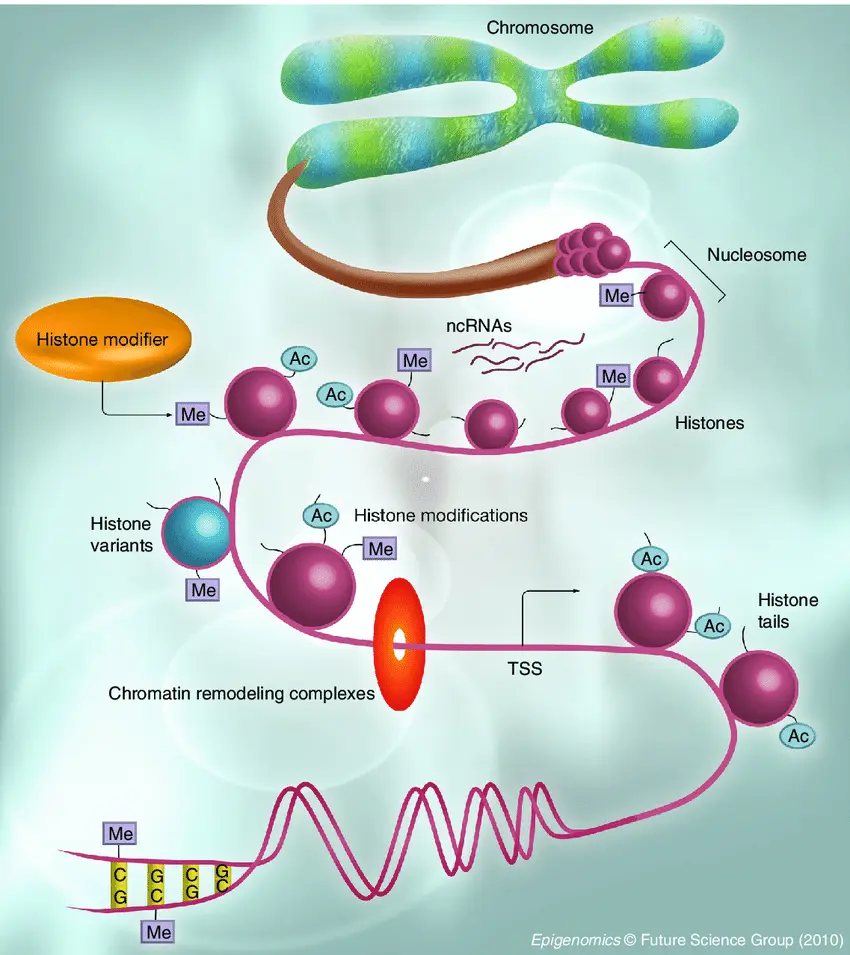
Epigenomic Sequencing
While traditional sequencing focuses on the genetic code itself, Epigenomic Sequencing explores chemical modifications to DNA and histones that regulate gene activity without changing the underlying sequence. These epigenetic marks control when and how genes are expressed. This technique helps researchers understand gene regulation during development, aging, stress responses, and disease processes like cancer, where epigenetic misregulation often plays a key role.
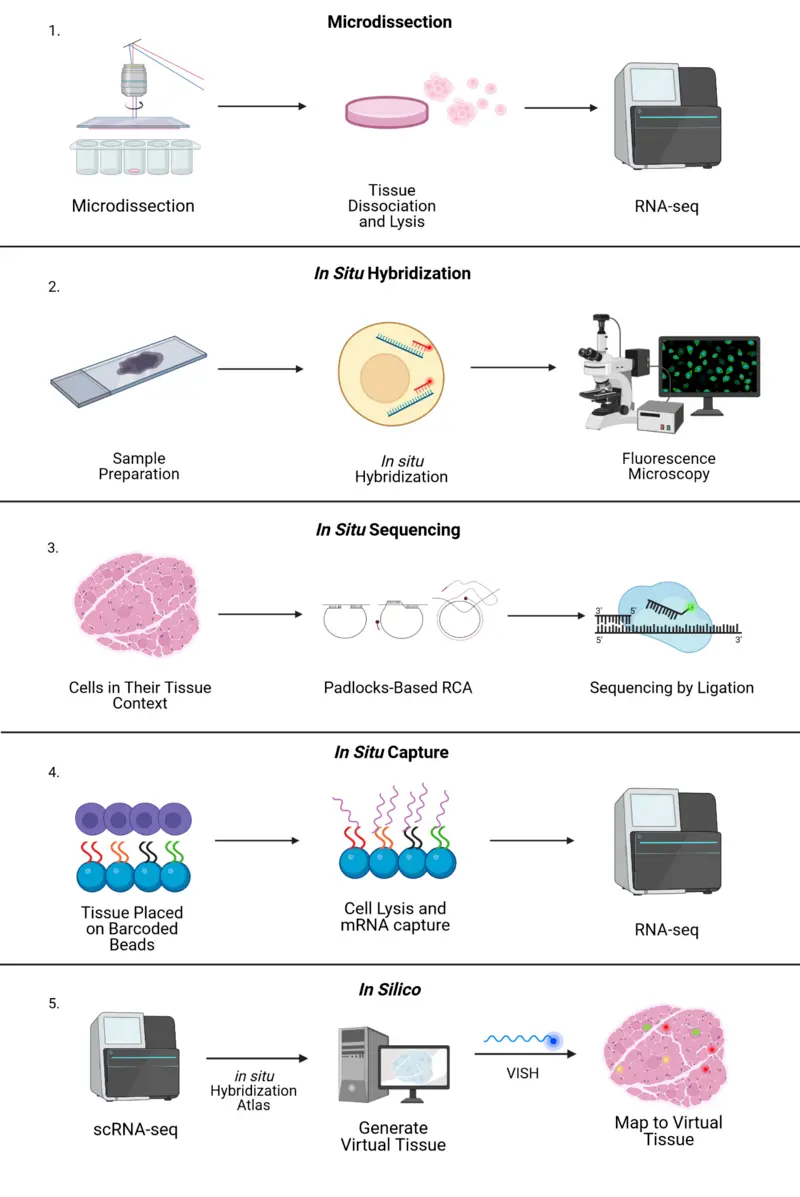
Single-Cell Sequencing
Single-Cell Sequencing allows the genetic or transcriptomic analysis of individual cells rather than bulk populations. This approach reveals important differences between cells that would otherwise be hidden in average measurements. Single-cell methods are essential for studying cancer heterogeneity, immune system function, and tissue development, as they uncover rare cell types and dynamic cellular changes that occur in complex biological systems.
Join the GCAT-SEEK Biotech Innovation community to explore the future of genomics, biotechnology, and Next-Generation Sequencing (NGS). Whether you are a researcher, educator, or biotechnology enthusiast, stay informed about the latest technologies, training workshops, and collaborative opportunities.